Introduction and demonstration
So you need to build a table model? 3ML via astromodels provides you with nice facilities to accomplish this task. But often, we need to interface with computationally expensive simulations and require many runs of these simulations. This is a very generic task and Ron Swanson wants to make things as simple as possible for you. Making things complicated is annoying.

Example
Let’s say we some simulation of an astrophysical spectral model that takes a very long time to compute for a given parameter set. We would like to fit this spectral model to some spectral data from an observatory, but each evaluation of the likelihood would be very costly. Instead, we can compute the simulation on some fixed grid of parameter values, and then interpolate between this grid when we need to perform our fit. This can dramatically decrease the evaluation time needed during the computation of the likelihood. To demonstrate, we will use a Band function (a type of smoothly broken power law) instead of an actual simulation, but the principle is the same as in the end a simualtion is a numerica function with input parameters and an outpu spectrum
We pick a parameter grid and a grid of energies for our simulation. We can enter these in a YAML file:
alpha:
custom: no
vmin: -1
vmax: 0
scale: "linear"
n_points: 10
beta:
custom: no
vmin: -3
vmax: -2
scale: "linear"
n_points: 10
epeak:
custom: yes
values: [50., 69.74 , 97.29, 135.72, 189.32, 264.097, 368.4, 513.9, 716.87, 1000.]
energy_grid:
custom: no
vmin: 10
vmax: 1000
scale: "log"
n_points: 50
As can be seen, we can specify the parameter/energy grids ourselves, or we can specify their ranges and let it be done for us.
It is possible that a simulation outputs more than one type of array (photons, electrons, neutrinos, etc.). In this case, each output may have its own energy grid. These can be specified as energy_grid_0, energy_grid_1…energy_grid_n. More on how to grab the output from these below.
The Simulation class
Now we need to make a class for the simulation. We will inherit from the simulation class and specify a _run_call function that tells the program how to run the simulation for a given set of parameters. This function must return a dictionary of arrays of photon / particle fluxes for the given energies. The keys of the dictionary should be output_0, output_1…output_n for each type of output corresponding to the energy grids above. If this were a call to compilcated
numerical simualtion, our call to the Band function would be replaced with that.
[1]:
from typing import Dict
import numpy as np
from astromodels import Band
import ronswanson as dukesilver
class BandSimulation(dukesilver.Simulation):
def __init__(
self,
simulation_id: int,
parameter_set: Dict[str, float],
energy_grid: np.ndarray,
out_file: str,
) -> None:
super().__init__(simulation_id, parameter_set, energy_grid, out_file)
def _run_call(self) -> np.ndarray:
# how parameters are translated into
# spectral output
b = Band(
K=1,
alpha=self._parameter_set["alpha"],
beta=self._parameter_set["beta"],
xp=self._parameter_set["epeak"],
)
return dict(output_0=b(self._energy_grid[0].grid))
16:17:23 WARNING The naima package is not available. Models that depend on it will not be functions.py:48 available
WARNING The GSL library or the pygsl wrapper cannot be loaded. Models that depend on it functions.py:69 will not be available.
WARNING The ebltable package is not available. Models that depend on it will not be absorption.py:33 available
WARNING We have set the min_value of K to 1e-99 because there was a postive transform parameter.py:704
WARNING We have set the min_value of K to 1e-99 because there was a postive transform parameter.py:704
WARNING We have set the min_value of K to 1e-99 because there was a postive transform parameter.py:704
WARNING We have set the min_value of K to 1e-99 because there was a postive transform parameter.py:704
WARNING We have set the min_value of F to 1e-99 because there was a postive transform parameter.py:704
WARNING We have set the min_value of K to 1e-99 because there was a postive transform parameter.py:704
The Simulation Builder
Now we need to tell the simulation builder a few things so it can construct our files for us. We have stored this YAML file in the repo itself. You should use your own!
The SimulationBuilder class takes a parameter grid, the name of the file that will be created, the import line for the custom simulation class, the number of cores and nodes to execute on.
We configure this with a YAML file.
# the import line to get our simulation
# we ALWAYS import as Simulation
import_line: "from ronswanson.band_simulation import BandSimulation as Simulation"
# the parameter grid
parameter_grid: "test_params.yml"
# name of our database
out_file: database.h5
# clean out the simulation directory after
# the run. It is defaulted to yes
clean: yes
simulation:
# number of multi-process jobs
n_mp_jobs: 8
[2]:
import ronswanson
sb = ronswanson.SimulationBuilder.from_yaml("sim_build.yml")
/opt/hostedtoolcache/Python/3.8.16/x64/lib/python3.8/site-packages/numba/core/ir_utils.py:2147: NumbaPendingDeprecationWarning:
Encountered the use of a type that is scheduled for deprecation: type 'reflected list' found for argument 'arrays' of function 'cartesian_jit'.
For more information visit https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-reflection-for-list-and-set-types
File "../../ronswanson/utils/cartesian_product.py", line 6:
@nb.njit(cache=True)
def cartesian_jit(arrays):
^
warnings.warn(NumbaPendingDeprecationWarning(msg, loc=loc))
However, you can easily do this from the command line:
> simulation_build sim_build.yml
Now a python file will be written to the disk which you can run to create your simulation runs. we can have a look at the file.
from ronswanson.band_simulation import BandSimulation as Simulation
from joblib import Parallel, delayed
from ronswanson import ParameterGrid
pg = ParameterGrid.from_yaml('/Users/jburgess/coding/projects/ronswanson/parameters.yml')
def func(i):
params = pg.at_index(i)
simulation = Simulation(i, params, pg.energy_grid.grid,'/Users/jburgess/coding/projects/ronswanson/database.h5')
simulation.run()
iteration = [i for i in range(0, pg.n_points)]
Parallel(n_jobs=8)(delayed(func)(i) for i in iteration)
Now this simply uses joblib to farm out the iterations over the parameter combinations. If iterations are to also be divided across HPC nodes, the python script will be modified and an associated SLURM script will be generated.
SLURM and advanced options
Configuring for SLURM and SBATCH systems is similar, but there are a few more options. ronswanson will set up bash scripts that will submit jobs to complete the simulations and then gather them into a database.
Here is an example script:
# the import line to get our simulation
# we ALWAYS import as Simulation
import_line: "from ronswanson.band_simulation import BandSimulation as Simulation"
# the parameter grid
parameter_grid: "test_params.yml"
# name of our database
out_file: database.h5
simulation:
# number of multi-process jobs PER node
n_mp_jobs: 9
# number of cpus to request per node (leave room for threads)
n_cores_per_node: 72
# number of runs per node
# here, the 9 mp jobs will take turns with these 500 runs
run_per_node: 500
# the switch to say we are performing SLURM job
use_nodes: yes
# optional maximum number of nodes to request
# if more than this are required, multiple
# submission scripts are generated
max_nodes: 300
# the max run time for each job in the array
time:
hrs: 10
min: 30
sec: 0
gather:
# after the simulations run
# you submit and MPI job that collects the simualtions
# number of simulations to collect per MPI rank
n_gather_per_core: 100
# number of cpus per node
n_cores_per_node: 70
# maximum job time
time:
hrs: 1
min: 0
sec: 0
Additional configuration of SLURM jobs can be handle with the ronswanson configuration.
The Database
Upon running the script, an HDF5 database of the runs is created which contains all the information needed to build a table model in 3ML.
[3]:
from ronswanson.utils.package_data import get_path_of_data_file
from ronswanson import Database
[4]:
db = Database.from_file(get_path_of_data_file("test_database.h5"))
db.parameter_ranges
db.plot_runtime()
Now we can use the database to construct and save a table model for 3ML
[5]:
table_model = db.to_3ml("my_model", "a new model for 3ML")
table_model
16:17:26 WARNING Energy unit is not a Quantity instance, so units has not been provided. template_model.py:106 Using keV.
[5]:
- description: a new model for 3ML
- formula: n.a.
- parameters:
- K:
- value: 1.0
- desc: None
- min_value: None
- max_value: None
- unit:
- is_normalization: False
- delta: 0.1
- free: True
- scale:
- value: 1.0
- desc: None
- min_value: None
- max_value: None
- unit:
- is_normalization: False
- delta: 0.1
- free: True
- alpha:
- value: 0.0
- desc: None
- min_value: -1.0
- max_value: 1.0
- unit:
- is_normalization: False
- delta: 0.1
- free: True
- beta:
- value: -2.5
- desc: None
- min_value: -3.0
- max_value: -2.0
- unit:
- is_normalization: False
- delta: 0.25
- free: True
- epeak:
- value: 226.70849609375
- desc: None
- min_value: 50.0
- max_value: 1000.0
- unit:
- is_normalization: False
- delta: 22.670849609375
- free: True
- K:
[6]:
import matplotlib.pyplot as plt
from astromodels import Band
%matplotlib inline
We can compare our table model with the Band function.
[7]:
import numpy as np
ene = np.geomspace(10, 1000, 100)
b = Band()
b.alpha = -0.6
b.beta = -2.5
b.xp = 250
b.K = 1.05
table_model.alpha = -0.6
table_model.beta = -2.5
table_model.epeak = 250
fig, ax = plt.subplots()
ax.loglog(
ene, ene**2 * table_model(ene), color='#33FFC4', lw=3, label="table"
)
ax.loglog(ene, ene**2 * b(ene), color='#C989FB', lw=3, label="band")
ax.set(xlabel="energy", ylabel="vFv")
ax.legend()
[7]:
<matplotlib.legend.Legend at 0x7f20283e1700>
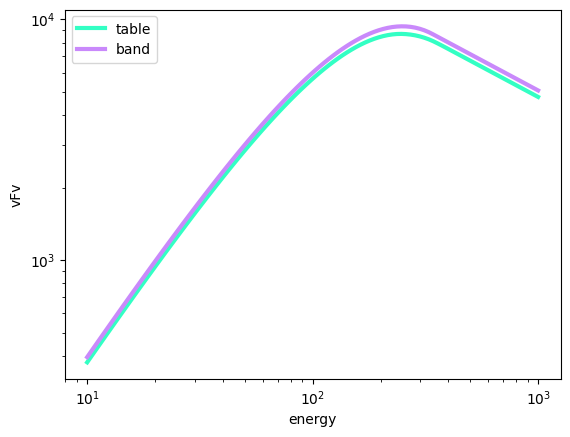
Great! That was way easier than programming everything yourself.

When you are ready to fit your model to data, one can follow the normal procedure implemented in 3ML.
Sub-selections
Suppose we did not want to use all the values in the parameter ranges we have simulated. Bigger interpolation tables take up memory when fitting.
We can select a subset of the parameter ranges when building the table
[8]:
selection = {}
selection['alpha'] = dict(vmax=0)
selection['epeak'] = dict(vmin=200, vmax=700)
table_model_small = db.to_3ml(
"my_model_small", "a new model for 3ML", **selection
)
table_model_small
16:17:29 WARNING Energy unit is not a Quantity instance, so units has not been provided. template_model.py:106 Using keV.
[8]:
- description: a new model for 3ML
- formula: n.a.
- parameters:
- K:
- value: 1.0
- desc: None
- min_value: None
- max_value: None
- unit:
- is_normalization: False
- delta: 0.1
- free: True
- scale:
- value: 1.0
- desc: None
- min_value: None
- max_value: None
- unit:
- is_normalization: False
- delta: 0.1
- free: True
- alpha:
- value: -0.5
- desc: None
- min_value: -1.0
- max_value: 0.0
- unit:
- is_normalization: False
- delta: 0.05
- free: True
- beta:
- value: -2.5
- desc: None
- min_value: -3.0
- max_value: -2.0
- unit:
- is_normalization: False
- delta: 0.25
- free: True
- epeak:
- value: 368.3999938964844
- desc: None
- min_value: 264.09698486328125
- max_value: 513.9000244140625
- unit:
- is_normalization: False
- delta: 36.83999938964844
- free: True
- K:
Awesome! Now go enjoy your weekend.

User configuration
A simple YAML configuration is stored in ~/.config/ronswanson/ronswanson_config.yml. It allows for configuring the log as well as putting default SLURM configuration parameters.
An example:
logging:
'on': on
level: INFO
slurm:
# where to send SLURM emails
user_email: my_email.com
# modules to be loaded for MPI jobs (gather script)
mpi_modules: ['intel/21.4.0', 'impi/2021.4',
'anaconda/3/2021.11','hdf5-mpi/1.12.2', 'mpi4py/3.0.3', 'h5py-mpi/2.10']
# modules to load for simulation jobs
modules: ['intel/21.4.0', 'impi/2021.4', 'anaconda/3/2021.11','hdf5-serial/1.12.2']
# the python binary for running the simulation jobs
python: "python3"
# where to store the simulations before
# database creation (default will be the directory where the code is run)
store_dir: /ptmp/jburgess
The configuration can be modified on the fly.
[9]:
from ronswanson import ronswanson_config, show_configuration
[10]:
show_configuration()
[10]:
config ┣━━ logging ┃ ┣━━ on: True ┃ ┗━━ level: WARNING ┗━━ slurm ┣━━ user_email: user@email.com ┣━━ modules: None ┣━━ mpi_modules: None ┣━━ python: python ┗━━ store_dir: None
[11]:
ronswanson_config.slurm.user_email
[11]:
'user@email.com'
[12]:
ronswanson_config.slurm.user_email = "workemail@email.com"
[13]:
show_configuration()
[13]:
config ┣━━ logging ┃ ┣━━ on: True ┃ ┗━━ level: WARNING ┗━━ slurm ┣━━ user_email: workemail@email.com ┣━━ modules: None ┣━━ mpi_modules: None ┣━━ python: python ┗━━ store_dir: None
[ ]: